05 May 2020
Meet the scientist - Dr Jason Roberts
Q&A with the Royal Melbourne Hospital’s Dr Jason Roberts, Head of the Electron Microscopy and Structural Virology Laboratory
What is your role at the Doherty Institute?
I’m currently responsible for the operational aspects of the Electron Microscopy Laboratory and the development and integration of advanced structural modelling techniques for diagnostic pathology, reference and research. Given my background in general virology, I also provide advice to other laboratories and government bodies regarding virology diagnostics and public health surveillance.

Dr Roberts’ day typically begins at 5:30am at the electron microscope (pictured here), ensuring the microscope is functioning and the environment stable for the day’s work to start.
What is an electron microscope and how does it differ from a normal microscope?
An electron microscope is very similar in principle to a normal light microscope. The main difference is that an electron microscope uses an electron beam rather than a beam of light to create an image of the specimen being examined. The use of electrons in an electron microscope allows the specimen to be magnified hundreds of thousands, even millions of times, compared to a conventional light microscope that can magnify an image a little over a thousand times. Additionally, the cost to purchase and install an entry level transmission electron microscope is measured in the millions of dollars, whereas a good quality light microscope costs almost a thousand times less.
What made you choose to specialise in the field of structural virology and emerging pathogens?
Before I started in science, I was a musician and enjoyed visual arts, so I think I’ve gravitated toward structural virology as it has allowed me to see the inherent beauty in the structures of the viruses we study. Prior to that, my work in diagnostic virology was more abstract; with the electron microscope I can actually see and model the viruses directly. Plus, the viruses look really cool when you import them into a virtual-reality headset.
With respect to emerging pathogens, I guess growing up I’ve always been terrified of viruses after watching movies like Outbreak. I have a tendency to study as much as I can about things that scare me to try to understand more about them. I’m also scared of sharks, so I spent years reading everything I could regarding sharks and did my advanced open water diving training so that I could go shark diving. It didn’t help - I’m still scared of sharks.
What projects are you working on at the moment?
Well I guess the most obvious one at the moment is a collaboration with the Victorian Infection Diseases Reference Laboratory (VIDRL) Virus Identification Laboratory and Bio21, visualising and performing 3-dimensional (3D) reconstruction of SARS-CoV-2, the virus that causes COVID-19. Before that, my primary research project was focussed on the application of advanced cryogenic electron microscopy techniques to examine emerging viral pathogens that require high containment.
My routine work relates to the visualisation of viruses associated with human pathology, with the priority being diagnostics. For example, polymerase chain reaction (PCR) methods are not always able to identify a particular pathogen straight away. So, if you grow a mystery bug in culture it makes life a lot easier if you can determine the morphology of the agent or determine ultrastructural changes made to the host cell. I also assist other labs within the Doherty Institute with their electron microscopy needs.
You’re an award-winning molecular modeller and scientific illustrator. How does 3-dimensional modelling support our understanding of viruses?
Determining the 3D structure of a virus, or specific components of a virus, can be very useful in understanding how the virus behaves and provides information that may be useful in the development of therapeutic interventions. When combined with supercomputer modelling, mechanisms of resistance can be hypothesised and further researched.
Perhaps the most powerful aspect of the work relates to the visual outputs, pictures, animations or 3D models that can assist in the education of students or helping with public engagement on complex topics. The saying “a picture is worth a thousand words” is very true.
An even more powerful tool that I have been working on is the integration of structural virology and virtual reality. I have witnessed members of the general public almost instantly grasping concepts such as antiviral binding and resistance mechanisms just by immersion in a 3D virtual reality environment. It’s fascinating to watch.

Dr Roberts’ images of the Novel Enterovirus A120 (left) and Novel Norovirus (right) were awarded gold at the 2018 Biennial Conference for the Australian Institute of Medical and Biological Illustrators
What process takes place behind the scenes to create a 3D reconstruction of a virus?
Unfortunately, the process is long and labour intensive at this point in time. To start the process, you need to have a very high concentration of good quality viruses. This is achieved by growing viruses in cell culture and purifying them. Ultra-clean, concentrated virus samples are critical to successful 3D reconstruction. Depending on the agent, this in itself can be quite problematic, especially with viruses that require high containment. In these instances, it’s crucial that the viruses are proven to be completely inert so they can be handled, but not significantly damaged so they can be observed in as close to a natural state as possible.
Once good quality virus preparations are available, the virions (or isolated proteins) are suspended in vitreous ice, a process that requires rapid freezing to -195 degrees Celcius almost instantaneously. After vitrification, the sample is held under liquid nitrogen until it can be loaded into the electron microscope in a special holder that maintains the temperature below -175 degrees Celcius.
We then use the electron microscope to take images of thousands or millions of viruses and use very powerful computers and complex programs to reconstruct the viruses (or proteins) in 3D. It is now possible to resolve 3D structures to the sorts of resolutions previously only attainable using highly specialised methods, such as x-ray diffraction.
What does a typical day look like in your role?
Electron microscopes are like formula one cars: they are expensive, operate at incredibly fine tolerances and are very temperamental. I typically start my day at 5:30am checking to make sure the microscope is functioning and the environment is stable enough to proceed with the day’s work. I’ll then begin cooling the microscope down with liquid nitrogen for at least an hour and then check other ancillary equipment to make sure there are no issues. While the microscope is cooling down, I tend to check email and perform administrative tasks.
Most of the day is spent juggling the preparation and imaging of samples with meetings and laboratory compliance work. Somewhere in between I try to write manuscripts, manuals and perform image analysis. There are never enough hours in the day, but I’m sure I’m not alone in thinking that.

Dr Roberts working on the electron microscope in the laboratory.
What’s a personal highlight from your work that you’re proud of?
That’s a really tough question. I’m never really proud of anything I’ve done as I always think that I could have done better, hindsight is always 20/20. I guess the best parts of my career have been the collaborations, whether it be the computer-based atomic reconstruction of poliovirus or sitting in WHO laboratory network meetings with other representatives from other countries sharing our experiences and knowledge for the common good.
I think that recently the most exciting thing has been collaborating with the VIDRL Virus Identification Laboratory to get the absolute best quality SARS-CoV-2 images we possibly could. It took a few days to tweak the methods but we have some amazing images of the virus.
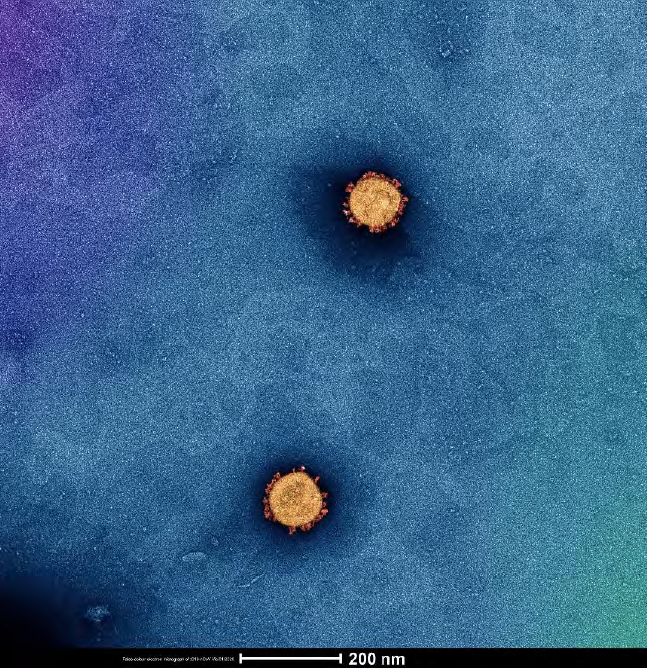
High quality image of SARS-CoV-2 captured on 7 February 2020 by Dr Roberts with technical assistance from Dr Andrew Leis.


