02 Jul 2018
Norovirus surveillance report for Victoria: January - June 2018
Dr Leesa Bruggink, Senior Medical Scientist with the Victorian Infectious Diseases Reference Laboratory, Royal Melbourne Hospital, at the Peter Doherty Institute for Infection and Immunity
Noroviruses are considered the leading cause of acute gastroenteritis, both sporadic and outbreak, in all age groups [1,2]. Noroviruses are classified into 6 established genogroups (GI – GVI), of which GI, GII, and GIV can infect humans [2]. Each norovirus genogroup can be further divided into genotypes, with the GII.4 genotype causing approximately 70-80% of all norovirus-associated gastroenteritis worldwide since the mid-1990s [3]. GII.4 noroviruses are particularly successful as they have been seen to evolve over time, through both antigenic drift and recombination, allowing the emergence of new variants that can evade a population’s herd immunity [3,4,5].
Faecal specimens from gastroenteritis outbreaks notified to the Victorian Department of Health and Human Services (DHHS) are sent to VIDRL for norovirus testing using real-time reverse transcription PCR (qRT-PCR). The first positive specimen from every norovirus positive outbreak then undergoes genotyping for epidemiological purposes. Where possible, genotyping is performed in both the open reading frame (ORF) 1 region (polymerase) and the ORF2 region (capsid) to ensure recombinant viruses are identified.
As at 29 June 2018, 189 specimens from 64 DHHS gastroenteritis outbreaks have been submitted to VIDRL for norovirus testing. Of these 64 outbreaks, 40 (62.5%) had norovirus detected in one or more specimens by norovirus qRT-PCR. The most common setting of norovirus positive outbreaks was aged care facilities (17/40; 42.5%), followed by childcare centres (11/40; 27.5%).
From January to June 2018 the level of norovirus positive outbreaks remained at a baseline level (Figure 1), whereas last year there was a peak in norovirus positive outbreaks in May to August (Figure 1). Genotyping data has shown that the 2017 winter peak in norovirus incidence was primarily due to the emergence of GII.4 recombinant GII.P4_NewOrleans_2009/GII.4_Sydney_2012 as an epidemic strain [6].
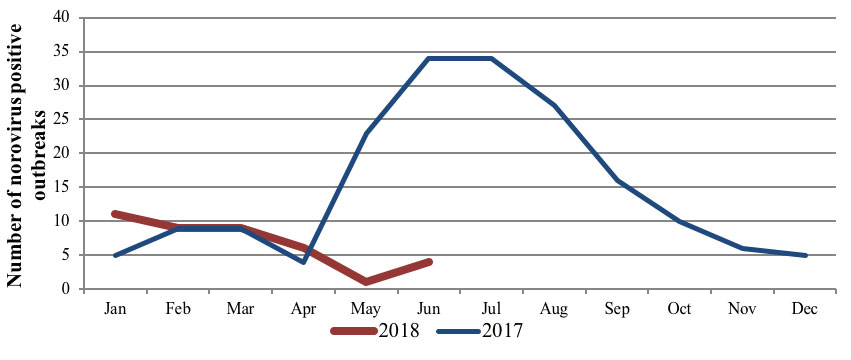
Figure 1: Number of norovrius positive outbreaks January - June 2018, compared to 2017
In contrast to 2017, genotyping data from 2018 (January to April; n=34) has shown the rise of a number of different genotypes. When ORF1 (polymerase) data is combined with the ORF2 (capsid) data, 24 outbreaks could be typed in both regions (Figure 2). The majority of the strains fully typed were recombinants (21/24; 87.5%), with the predominant strain being GII.P16/GII.2 (8/24; 33.3%). Interestingly, the epidemic GII.4 recombinant strain from 2017 has not been detected so far in 2018.
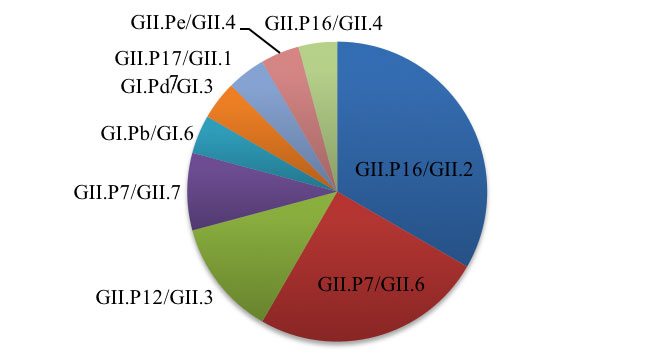
Figure 2: ORF1/ORF2 norovirus genotypes from outbreaks Jan-Apr 2018 (n=24)
The predominant strain in Victoria in 2018 (GII.P16/GII.2) has been associated with large increases of norovirus incidence in a number of countries around the world, including China in 2017 [7], France in the winter of 2016/17 [8], Germany in late 2016 [9], Italy in 2017 [10], and Japan in 2016/17 [11]. As surveillance data shows that this strain is now circulating at relatively high levels in Victoria, it could potentially be a primary contributor to the seasonal peak in norovirus incidence expected in the latter half of 2018.


